HBond Score
HEXplorer score
ModCon
HBond Score background
HBond Score Concept
Within eukaryotic cells, the formation of an RNA duplex between the free 5' end of the U1 snRNA and its target sequence the pre-mRNA stimulates the formation of the spliceosome that catalyzes splicing. Sequence compilation of thousands of human 5' splice sites (Fig. 1) reveal a so-called consensus sequence, i.e., AG/GURAGU (where R=purine, and / indicates the exon-intron border) that reflects the frequency of a nucleotide in a given position of such a compilation. However, the biological significance of this consensus sequence remains arguable as numerous approaches which have been suggested for the prediction of 5' splice sites exclusively based on, e.g., nucleotide consensus matrices, fail to be highly reliable.
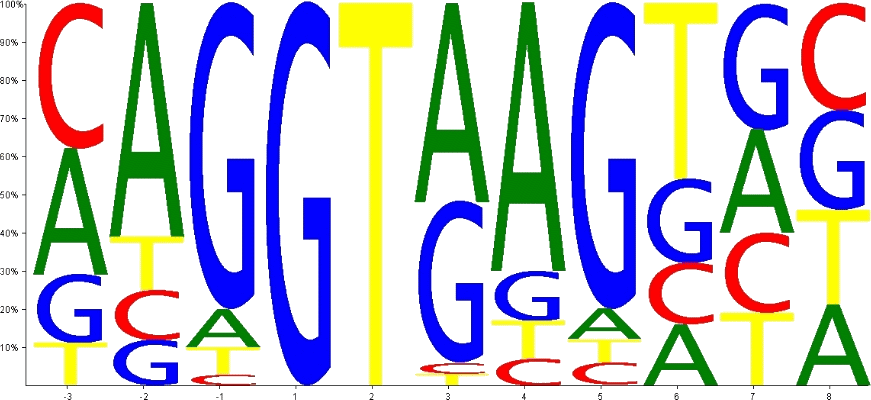
Figure 1 Sequence logo of 27,054 human 5' splice sites.
An obvious limitation of the current 5' splice sites consensus sequence becomes most evident for two functional U1 snRNA binding sites that we have analyzed recently, and which had only three nucleotides in common. Nevertheless, despite their sequence divergence both 5' splice sites could form an RNA duplex with the U1 snRNA of 14 hydrogen bonds suggesting to us that a significant consensus might be based on hydrogen bonding rather than on the sequence (Fig. 2).

Figure 2: Immunoblot analysis (left) of glycoproteins expressed in HeLa-T4+ cells transfected with SVcrev, pGL3-control and env expression vectors carrying predicted U1 snRNA binding sites in the position of the tat/rev 5' splice site SD4. The hydrogen bonding pattern is calculated on the basis of the sequence complementarity between the 5' end of U1 snRNA and the 5' splice site. The numbers represent the numbers of H-bonds in a continous stretch of complementary nucleotides. A vertical bar indicates a mismatch. If env expression is detectable the algorithm assesses the hydrogen bonding pattern as HC (high complementarity), if no env expression is detectable as LC (low complementarity) the threshold value.
Therefore, based on a mutational analysis, we developed a new algorithm that analyses individual hydrogen bonding patterns of a U1 snRNA binding site, irrespective of any nucleotide frequency. Experimentally a threshold level is defined that is of predictive value for identifying U1 snRNA binding sites downstream of the HIV-1 exonic splicing enhancer of the second rev exon. Our model postulates that different binding patterns of SR proteins determine different threshold values for U1 snRNP binding (Fig. 3).

Figure 3: An SR protein recruits U1 snRNP to the 5' splice site. The number of complementary nucleotides necessary for U1 snRNA binding can vary between different SR proteins. In (A) the SR protein supports U1 snRNA binding via interaction with the U1 70K stronger than in (B). Thus, the threshold value for the 5' splice site in the context of (A) is lower than in (B).
References
Corinna Asang, Ilona Hauber, Heiner Schaal. 2008. Insights into the selective activation of alternatively used splice acceptors by the human immunodeficiency virus type-1 bidirectional splicing enhancer. Nucleic Acids Res., 1-14;
Marcel Freund, Martin J. Hicks, Carolin Konermann, Marianne Otte, Klemens J. Hertel, Heiner Schaal. 2005. Extended base pair complementarity between U1 snRNA and the 5' splice site does not inhibit splicing in higher eukaryotes, but rather increases 5' splice site recognition. Nucleic Acids Res., 33 (16):5112-5119;
Caputi M, Freund M, Kammler S, Asang C, Schaal H. A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression. J Virol. 2004 Jun;78(12):6517-26.
Freund, M., C. Asang, S. Kammler, C. Konermann, J. Krummheuer, M. Hipp, I. Meyer, W. Gierling, S. Theiss, T. Preuss, D. Schindler, J. Kjems, and H. Schaal. 2003. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res., 31:6963-6975
Kammler, S., C. Leurs, M. Freund, J. Krummheuer, K. Seidel, T. Tange, J. Kjems, A. Scheid, and H. Schaal. 2001. The sequence complementarity between HIV-1 5' splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA, 7:421-434